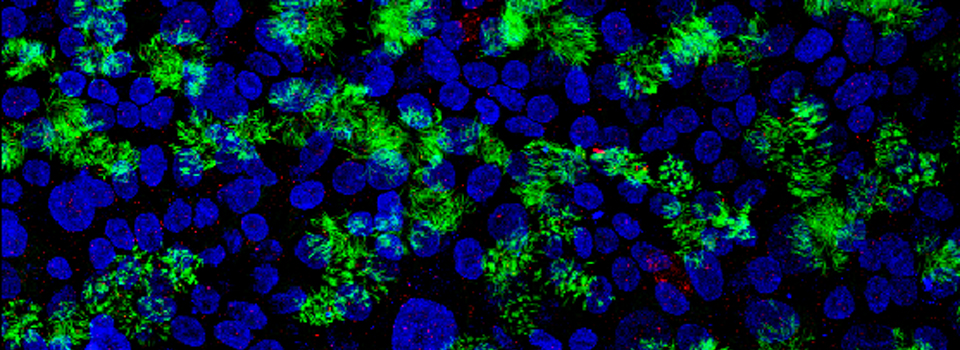
Prepping image data for FARSIGHT
For most imaging experiments done on the confocal microscopes, a user wants to assemble all the individual XY frames into a larger, combined image and/or movie. If you want to run your data through the FARSIGHT program, you will need all those separate individual frames in TIF format.
The example below shows a tile scan taken with the Leica SP8.
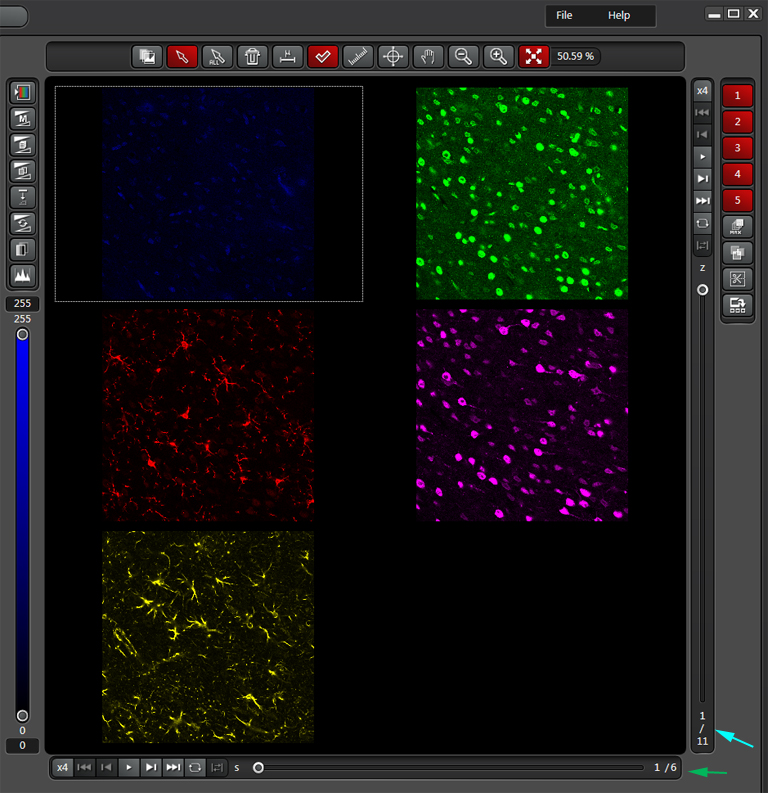
It is a 3 x 2 mosaic with 6 stacks of 11 z slices each, in 5 different channels. The display is set to show stack 1 of 6 stacks (slider at the bottom, green arrow) and Z slice 1 of 11 slices (slide at right, cyan arrow). Therefore the total raw data set is comprised of 6 stacks x 11 z-slices x 5 channels = 330 individual xy images. To generate all those images, right click on the raw data file, and choose “export”, then “TIF”. This will open an “Export to TIFF” window. Make sure that the “Save RAW Data” box is the only one checked. Use the “Browse” button to select the destination folder, then click “Save”.
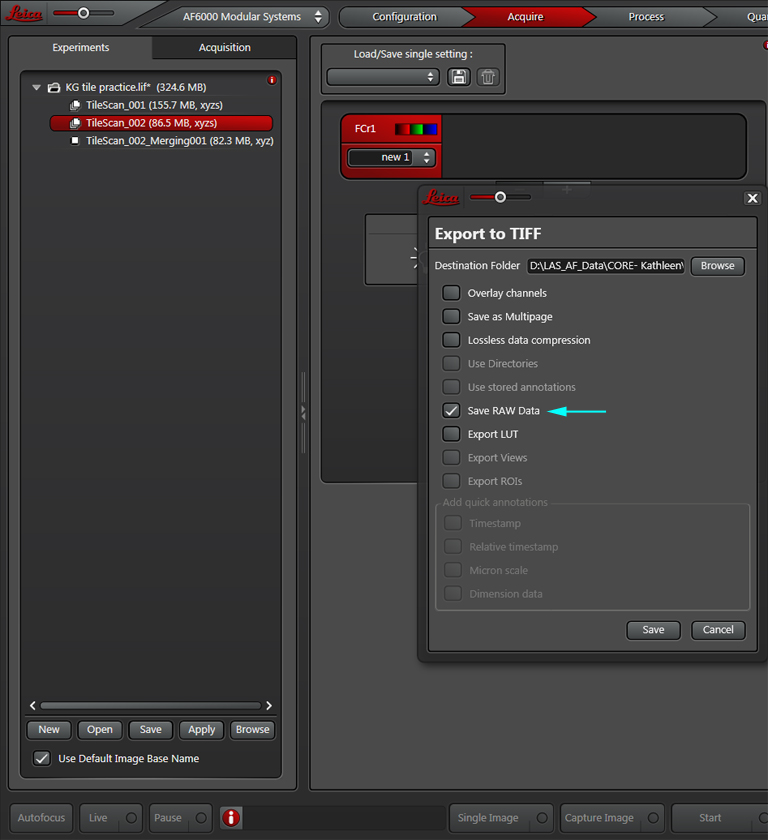
This will dump all the TIF files into your destination folder. Next you will need to organize them for loading into Farsight. You do this by grouping them first by channel, and then by stack within each channel folder.
Each TIF file is named according to its position in the combined data set.
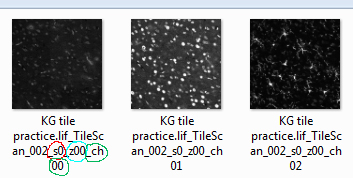
One quirk of this system is that it starts numbering from “0”, rather than “1”, so you must keep that in mind as you organize the files. The relevant parts are the stack # (s, circled in red), the z position (z, circled in cyan), and the channel # (ch, circled in green). The icon on the left represents the first z slice of stack #1 in channel 1, the next image the first z slice of stack #1 in channel 2, etc.
The search function of the workstation PC is useful for isolating groups of files by their channel. In this example, to isolate all files from channel 1 (which is named”ch00”), type “ch00 tif’ into the search function of the window:
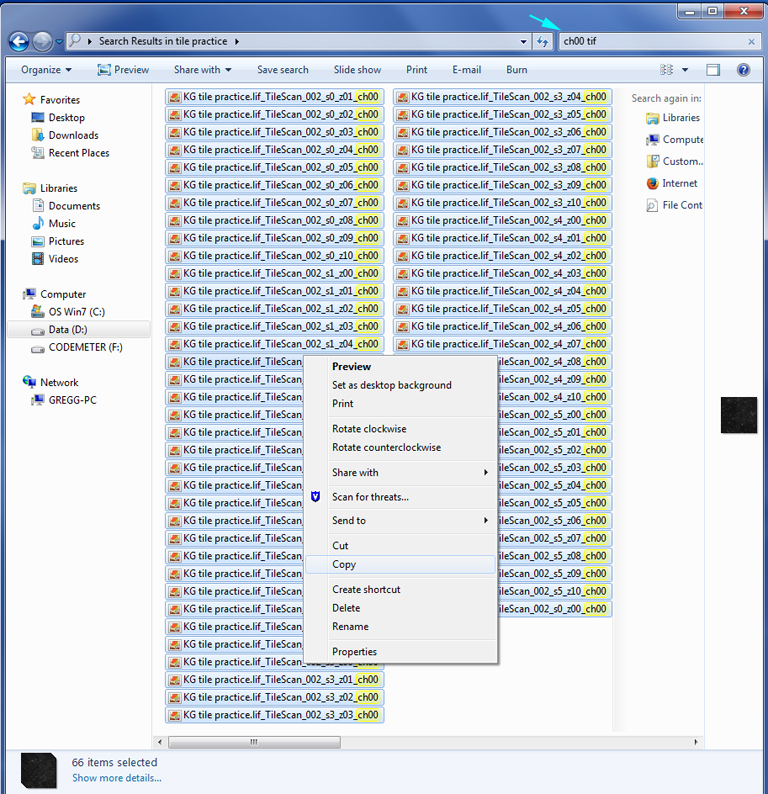
This will separate out all the tif files for “ch00” (channel 1). Copy those files into a new folder.
Next you will need to separate all these channel 1 files by stack. Create a new folder (Ch1 stack 1), select all the files with “s0”, and move then to the new folder. Repeat this for each stack.
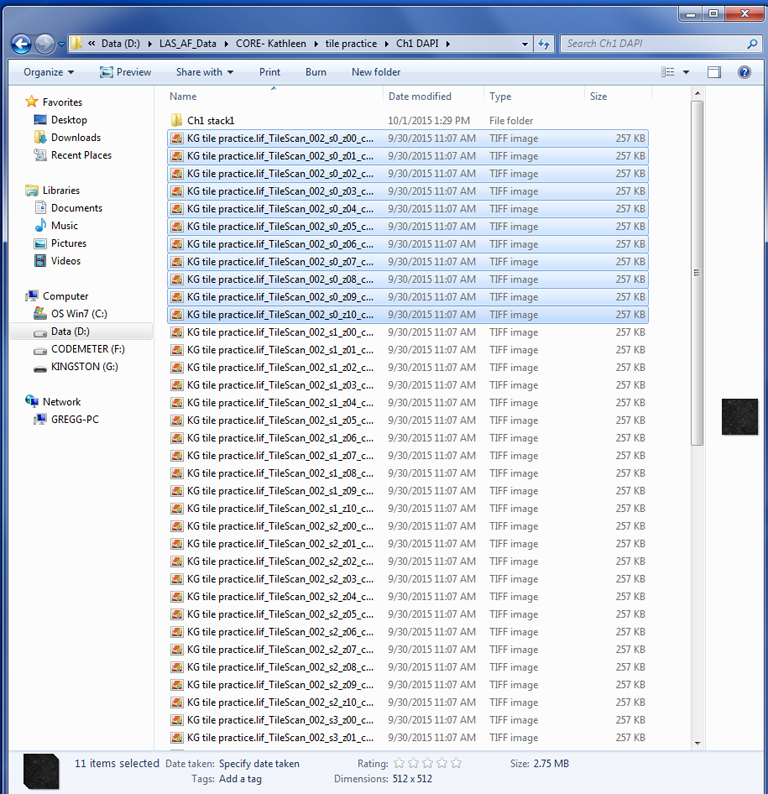
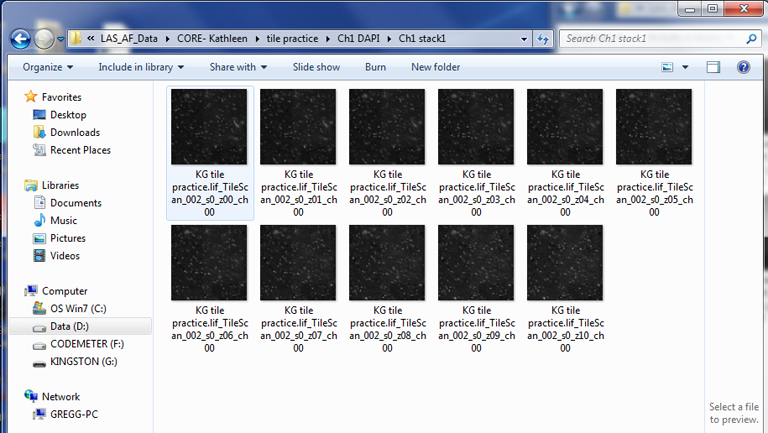
Each stack folder should have the complete Z-dimension series for that stack/ channel:
When you are finished sorting the tiffs for the channel, you should have a folder of Z-slices for each stack:
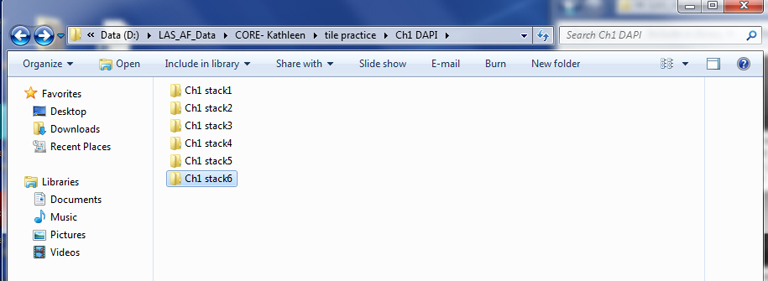
Repeat this for all channels.
The final step is to rename the individual files to remove any characters that are incompatible with the FARSIGHT software, such as any dots or spaces. Unfortunately the Leica software does include “.lif” in the file names, so you should remove all the “.” before starting any analysis of the files.